 Use case title (contact person)
Use case title (contact person)
SPELMED: Population Genomics analysis of the small pelagic fish stocks in the Northwestern Mediterranean Sea (Dr. Costas Tsigenopoulos)
In-brief: description the analysis that was carried out
ddRAD data were sequenced for anchovy and sardine populations across the Mediterranean. 6 HiSeq lanes produced 2 billion Illumina reads which were used for SNP discovery and genotyping.
In brief: description of the pertinent project
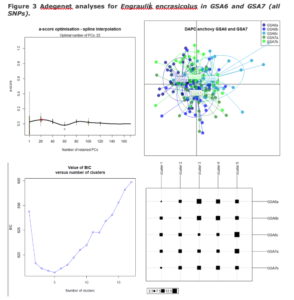
We document the extent of genetic differentiation within and among geographic sampling locations of the European sardine (Sardina pilchardus) and the European anchovy (Engraulis encrasicolus) mainly from W. Mediterranean locations as well as adjacent Atlantic ones. Using a genome-wide data set of a great dataset of double-digest RAD-derived SNPs, we show that the most important component of genetic differentiation among populations is found between the Atlantic and the W. Mediterranean whereas the Alboran Sea samples are also of distinct status in both species.
Comments on how the cluster supported this work
In total 6 nodes were used for two months including various software such as STACKS, STRUCTURE, fastSTRUCTURE, adegenet.

