 Use case title (contact person)
Use case title (contact person)
Genome analysis of Pseudomonas strains isolated from Kolumbo, Santorini (Panos Bravakos).
In-brief: description the analysis that was carried out
Genome assembly, genome annotation, pangenome, phylogenomics
In brief: description of the pertinent project
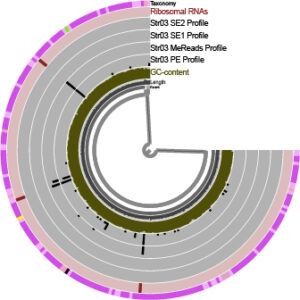
23 non-axenic isolated strains of the genus Pseudomonas were sequenced in Illumina Miseq. Reads were filtered based on their quality, assembled into scaffolds, and subsequently annotated by various databases. All genes from the 23 genomes were clustered together to form the pangenome. Genomes from the genus Pseudomonas were downloaded from public databases to construct a maximum likelihood phylogenomic tree.
Comments on how the cluster supported this work
A series of bioinformatic tools were used like Fastp, Spades, Anvi’o, RaxML and in-house scripts. Many hours of cpu time usually in batch.

