 Use case title (contact person)
Use case title (contact person)
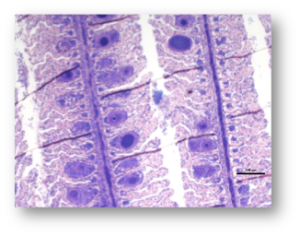
Hybrid assembly and genome analysis of an epitheliocystis related agent. (Maria Chiara Cascarano)
In-brief: description the analysis that was carried out
Six samples containing pooled cysts were sequenced both with MinION and Miseq. Long and short reads were demultiplexed, quality filtered and trimmed. Six metagenome assemblies were than built with different softwares using long reads and subsequently polished using short reads. Assembly quality statistics was performed to compare results obtained from different tools. Binning of the metagenomic contigs allouded the separation of the clusters containing the bacterial genome.
In brief: description of the pertinent project
Epitheliocystis has been reported in different fish species and several areas worldwide. This pathology is characterised by the presence of cyst-like inclusions on the epithelial surface of the gills containing the replicating infectious agent. The intracellular lifestyle of this bacteria implies the impossibility of isolating and culture this pathogens. Draft genomes have therefore to be obtained from direct sequencing of infected material (mini-metagenomes).
The project aim is to get insights on the infection mechanism by describing genomic features of the major epitheliocystis agent affecting Greater Amberjack in in Crete (Greece).
Comments on how the cluster supported this work
Software request and troubleshooting was supported by the cluster with tools as adapterremoval, trimmomatic, scytle, deepbinner, bwa, graphmap, canu, flye, pilon, Spades, Quast, busco, kraken, maxbin, concoct)
Resources: One node on batch partition discontinuously in different months.

