 Use case title (contact person)
Use case title (contact person)
Phylogenomics investigation of sparids (Teleostei: Spariformes) using high-quality proteomes highlights the importance of taxon sampling (Dr. Tereza Manousaki)
In-brief: description the analysis that was carried out
Phylogenomic pipeline including orthology analysis, multiple sequence alignment and tree inference
In brief: description of the pertinent project
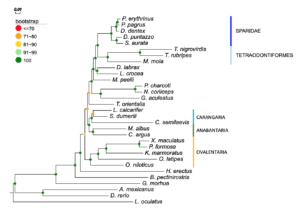
we revisited the phylogenetic relationship of sparids using newly obtained data from five genera with various reproductive modes, protogynous (Pagrus, Pagellus, Fig. 1A), protandrous (Sparus, Diplodus) and gonochoristic (Dentex). We built a phylogenomic dataset by including 26 non-sparid fish from a wide classification spectrum, focusing on high-quality, publicly available proteomes from species with sequenced genome, to ensure the highest data; uality possible. The inferred phylogeny proposed Tetraodontiformes as closest relative to sparids than all teleost groups with a sequenced genome, a robust finding well supported by a variety of applied phylogenetic tests.
Comments on how the cluster supported this work
In total 10 nodes were used for the orthology and phylogenomic Maximum likelihood analysis for multiple months. Multiple software were used such as RaxML Orthofinder, Orthomcl, etc.

