 Use case title (contact person)
Use case title (contact person)
SPARCOMP: Comparative Transcriptomics in Sparids brain and gonads shed light into sex differentiation (Dr. Costas Tsigenopoulos)
In-brief: description the analysis that was carried out
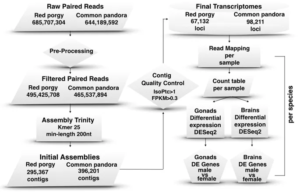
RNASeq analysis including raw read preprocessing, de novo transcriptome assembly, mapping, expression quantification and differential expression analyses.
In brief: description of the pertinent project
We sequenced and analyzed the sex-biased transcriptome in gonads and brain (the tissues with the most profound role in sexual development and reproduction) of two sparids with different reproductive modes. Through comparative analysis we put forward common male and female-specific genes and pathways that are probably implicated in sex-maintenance in this fish family. Our results contribute to the understanding of the complex processes behind the establishment of the functional sex, especially in hermaphrodite species and set the groundwork for future experiments by providing a gene toolkit that can improve efforts to control phenotypic sex in finfish in the ever-increasingly important field of aquaculture.
Comments on how the cluster supported this work
In total 6 nodes were used for two months including the bigmem node for the assembly. Multiple software were used such as Trimmomatic, Trinity, DESeq2, EdgeR, etc.

